Variants
There are a total of unique hgvs strings directly linked to individuals in this SQLite database. There are a total of variants with a ClinVar ID.
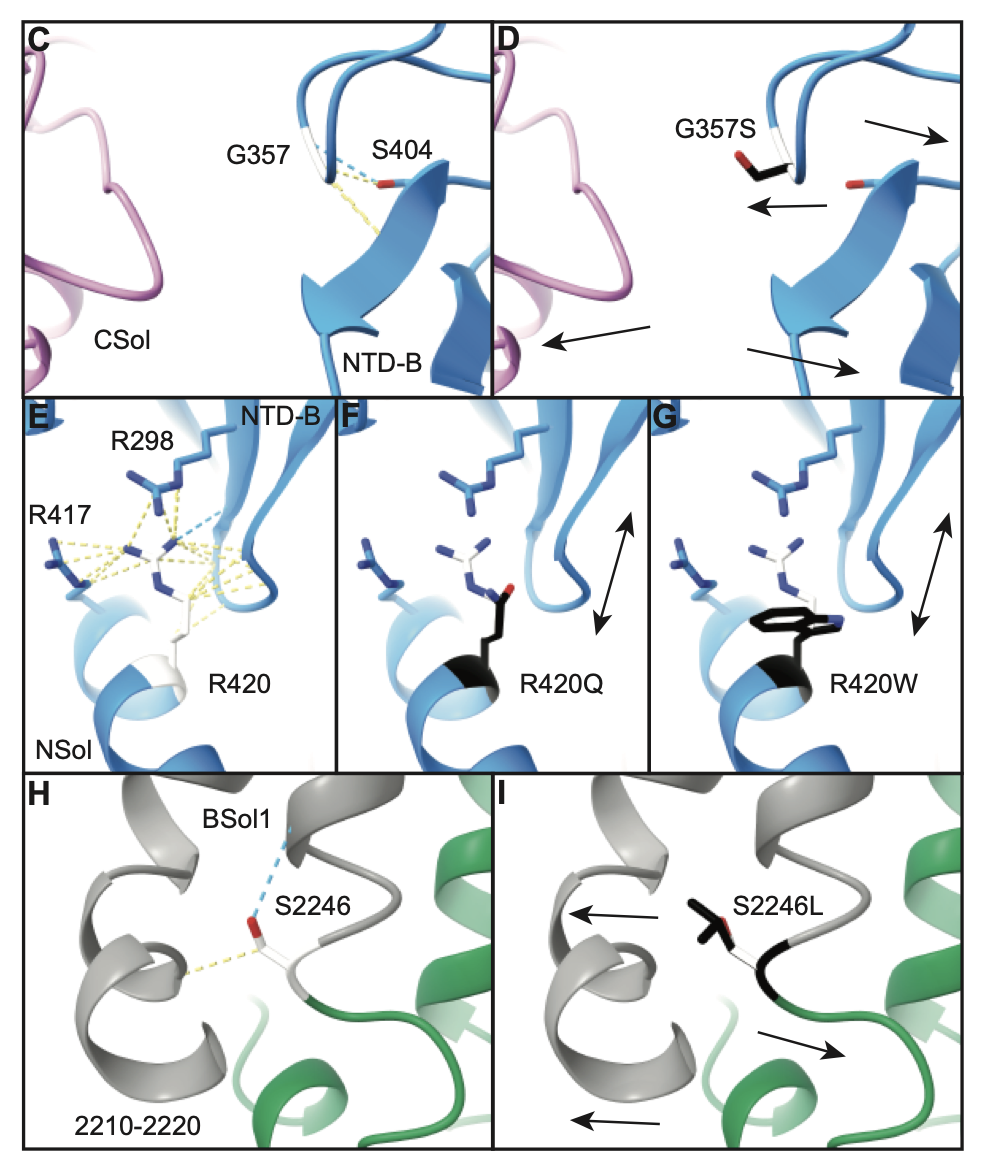
Gly357Ser (G357S), Arg420Gln (R420Q), Arg420Trp (R420W), and Ser2246Leu (S2246L) are the most common variants linked with patients in this database.
R420Q and R420W
Our lab has also looked at the effects of the R420Q and R420W variants and the effect on the structure of the RYR2 protein.
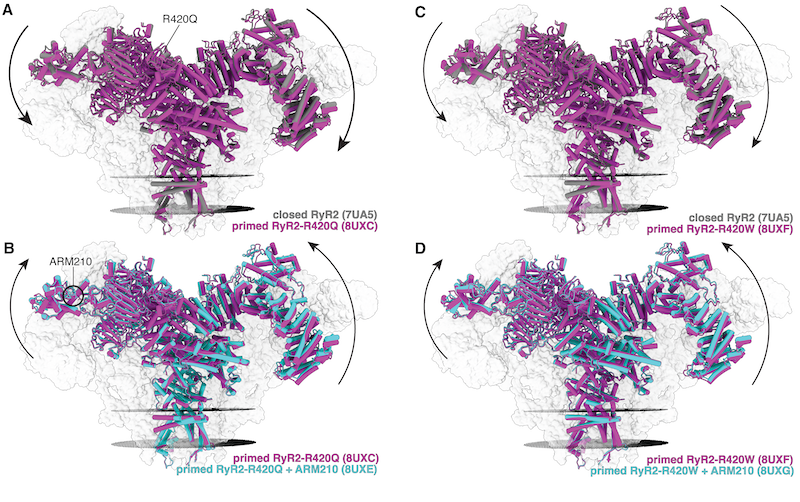
We can see from this figure above that the R420Q and R420W variants results in a shift in the conformation of the RYR2 protein.
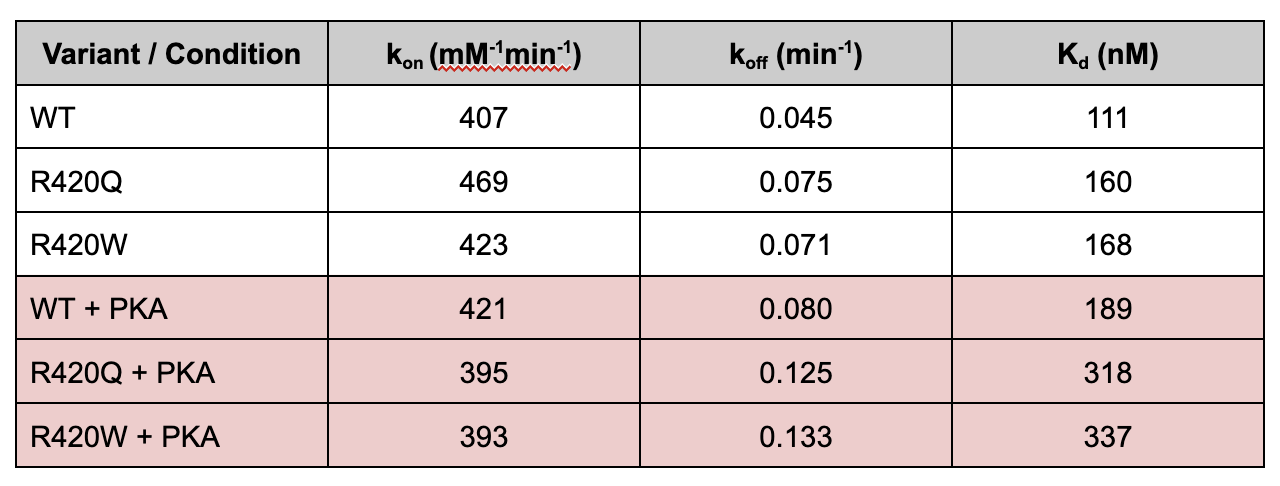
This shift decreases calstabin binding affinity of the R420Q and R420W variants. In the figure below, we can see that the R420Q and R420W has a significantly increased constant of unbinding compared to the wild type RYR2 protein, in the presence of protein kinase A (PKA). This results in a higher constant of dissociation, which means that the R420Q and R420W variants have a lower binding affinity for calstabin.
This shift ultimately results in a channel which is more likely to open because of the depleted calstabin - resulting in the calcium leak and the conditions associated with CPVT.
Location of these variants in the RyR2 protein
Using the @rcsb/rcsb-saguaro-3d package, we can visualize the location of
these
variants in the RyR2 protein structure.
Eventually, this sequence viewer below will take the user to the specific location within the 3D protein structure.
Recently, our lab has solved the structure of the RyR2 protein with the R420Q and R420W variants using cryo-EM.
Showing PDB:
Image of the variants in the RyR2 protein structure.
Exon location of variants
Using this database, we can also calculate the exon location of the variants. With this information, we can see if there are:
- exons that are more likely to have variants
- exons with a higher number of individuals with variants
- exons with a lower average age of onset compared to other exons.
We can see that variants are not evenly distributed, with many variants located in exons 8, 14, 47, 90. This matches up with previous literature, by Kapplinger et al.’s, which define many hotspots in the RyR2 gene with CPVT variants.
Average Age of Onset of CPVT per exon
SQL Query
Click to expand
SELECT (SELECT exon
FROM exon_in_coding_region_view
WHERE v.c_start_i BETWEEN nucleotide_start AND nucleotide_end) AS exon,
ROUND(AVG(age_of_onset), 2) AS avg_age_of_onset
FROM variant v
JOIN individual i
ON v.variant_id = i.variant_id
JOIN individual_condition ic
ON i.individual_id = ic.individual_id
JOIN condition c
ON ic.condition_id = c.condition_id
WHERE c.condition = 'Catecholaminergic polymorphic ventricular tachycardia 1'
AND exon IS NOT NULL
GROUP BY exon;
Using this data we can also see that there are exons with a lower average age of onset compared to other exons. This could be due to the location of the variants in the protein, and the effect on the protein structure.
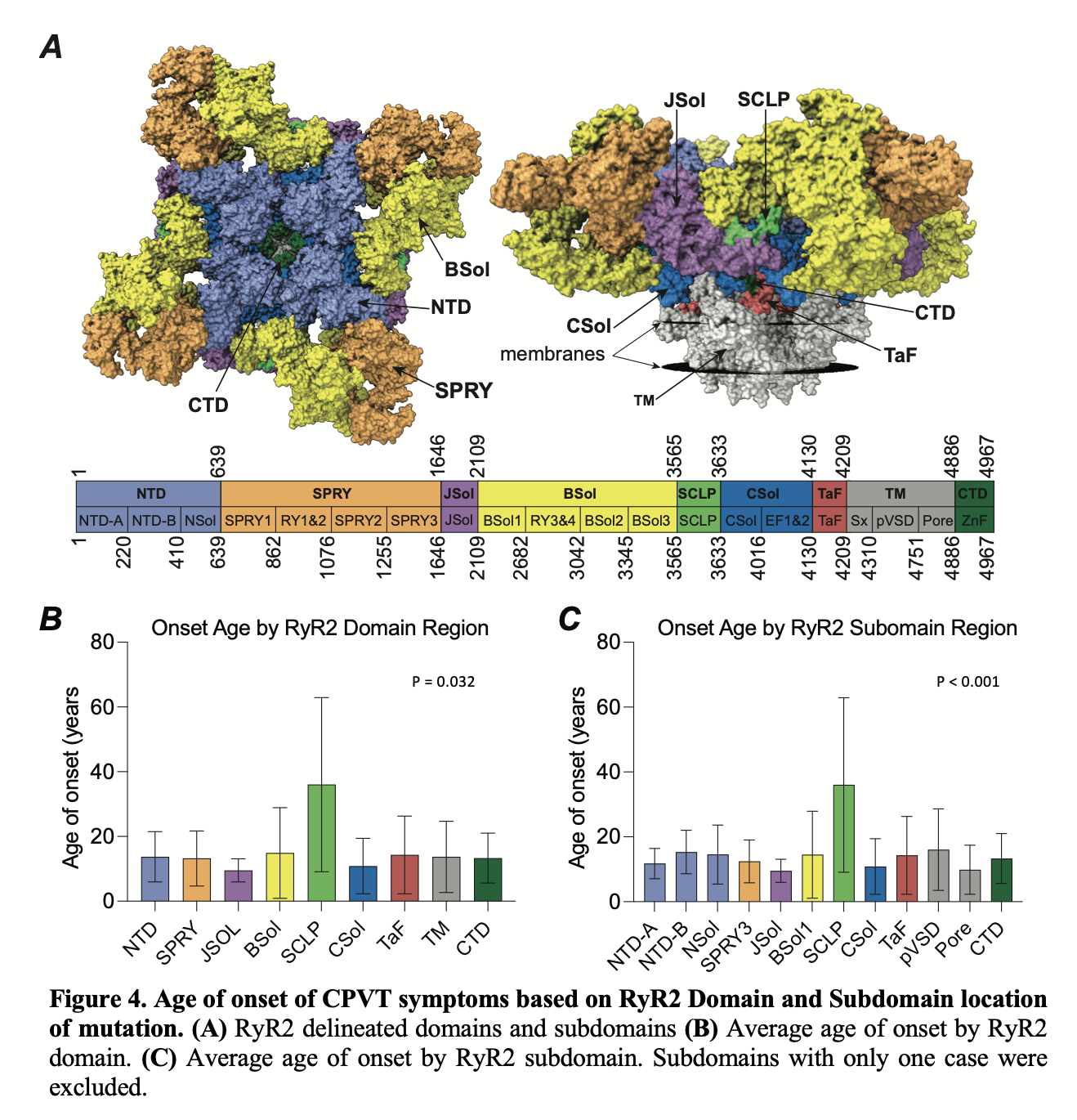
In the image below, we can see that the average age of onset of CPVT is different depending on the domain and subdomain of the RyR2 protein.
For example, variants in the JSOL domain have a significantly lower (p = 0.038) compared to variants located in the Shell-core linker peptide (SCLP) domain.